| 24. Regulating protein localization and activity | ||
|
|
|
|
Telling proteins where to go: Translation of proteins occurs in the cytoplasm, where mature ribosomes are located. If no information is added, a newly synthesized polypeptide will remain in the cytoplasm, that is its default location. |
| Yet even in the structurally simplest of cells, the bacteria and archaea, there is more than one place that a protein may end up: it can remain in the cytoplasm, it can be inserted into the plasma membrane or it may be secreted from the cell. Both membrane and secreted polypeptides must be inserted into, or pass through, the plasma membrane. |
|
Polypeptides destined for the membrane or for secretion are generally marked by a specific tag, known as a signal sequence. The signal sequence consists of a stretch of hydrophobic amino acids, often at the N-terminus of the polypeptide. As the signal sequence emerges from the ribosome it interacts with a signal recognition particle or SRP - a complex of polypeptides and a structural RNA. The binding of SRP to the signal sequence causes translation to pause. SRP acts as a chaperone for membrane proteins. |
|
The mRNA/ribosome/nascent polypeptide/SRP complex will find (by diffusion), and attach to, a ribosome/SRP receptor complex on the cytoplasmic surface of the plasma membrane (in bacteria and archaea.) This ribosome/SRP receptor is associated with a polypeptide pore. When the ribosome/SRP complex docks with the receptor, translation resumes and the nascent polypeptide passes through a protein pore and so through the membrane. As the polypeptide emerges on the extracytoplasmic side of the membrane, the signal sequence is generally removed by an enzyme, signal sequence peptidase. If the polypeptide is a membrane protein, it will remain within the membrane. If it is a secreted polypeptide, it will be released into the periplasmic space. |
|
Eukaryotic cells are structurally and topologically more complex than bacterial and archaeal cells; there are more places for a newly synthesized protein to end up. While we will not discuss the details of that process, one rule of thumb is worth keeping in mind. Generally, in the absence of added information, a newly synthesized polypeptide will end up in the cytoplasm. |
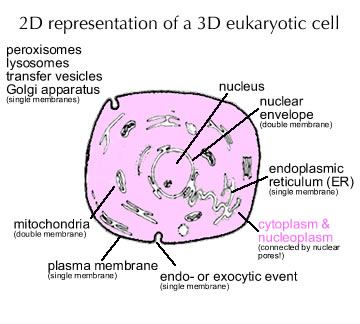 |
|
As in bacteria and archaea, polypeptides destined for secretion or insertion into internal membrane systems (that is the endoplasmic reticulum) are directed to their final location by the signal sequence/SRP system. Proteins that must function in the nucleus generally get there because they have a nuclear localization sequence or NLS. On the other hand, proteins can be excluded from the nucleus using a nuclear exclusion sequence or NES. Likewise, other localization signals and sequences are used to direct proteins to other intracellular compartments. |
|
|
|
Regulating protein activity: Proteins act through their interactions with other molecules. Enzymes interact with substrate molecules; these interactions lower the activation energy of the reaction's rate limiting step, leading to an increase in the overall reaction rate. At the same time, cells and organisms are not static. They must regulate protein activity to adapt to changes in their environment or within the organism. A protein's activity can be regulated in a number of ways. The first and most obvious is to control the total number of protein molecules present within the system. Let us assume that once synthesized, a protein is fully active. With this simplifying assumption, the total concentration protein in a system [Psys] is proportional to (the rate of protein synthesis) minus (the rate of protein degradation). What controls the rate of protein synthesis? Clearly this is primarily a function of the number of mRNAs encoding the protein present in the cell. It is generally the case that once translation begins, it continues at a more or less constant rate. |
|
In the bacterium E. coli, the rate of translation at 37ºC is about 15 amino acids per second. The translation of a polypeptide of 1500 amino acids therefore takes about 100 seconds. After translation, folding and, in multisubunit proteins, assembly, the protein will function until it is degraded. Protein degradation often begins when the protein is specifically marked for degradation. This is an active process, involving ATP hydrolysis and a multisubunit complex known as the proteosome (see previous page). The proteosome degrades the polypeptide into small peptides and amino acids, that can be recycled. Degradation has a drawback, however, it is irreversible. |
 |
|
A second and reversible form of regulation is known as allosteric regulation. In allosteric regulation, regulatory molecules bind reversibly to the protein, altering its confirmation, which in turn alters the protein's activity. Such allosteric effectors are not covalently attached to the protein. Here, the activity of a protein is positively regulated by the binding of a factor. |
|
This factor could be a small molecule or another protein. What is important is that the allosteric binding site is distinct from the enzyme's catalytic site. That is what allosteric means, other site. An allosteric effector can also act negatively, inhibiting an enzyme's activity. Because allosteric regulators do not bind to the same site on the protein as the substrate, changing substrate concentration generally does not alter their effects. |
|
Of course there are other types of regulation as well. Active site inhibitors: An inhibitory factor may bind to and block the active site. If this binding is reversible, then increasing the amount of substrate can over-come the inhibition. An inhibitor of this type is known as a competitive inhibitor. In some cases, the inhibitor chemically reacts with the enzyme, forming a covalent bond. Because this type of inhibitor is essentially irreversible, increasing substrate concentration does not overcome inhibition. These are therefore known as non-competitive inhibitors. Allosteric effectors are also non-competitive, since they do not compete with substrate for binding to the active site. That said, binding of substrate could, in theory, change the affinity of the protein for allosteric effectors, just as binding of the allosteric effector changes the binding affinity of the protein for the substrate. |
|
Post-translational regulation: Proteins may be modified after synthesis - this process is known as post-translational modification. A number of post-translational modifications have been found to occur within cells. The first type involve the covalent addition of specific groups to specific sites on the protein - these groups can range from phosphate groups (phosphorylation), an acetate group (acetylation), the attachment of lipid/hydrophobic groups (lipid modification), or carbohydrates (glycosylation). Many post-translational modifications are reversible, one enzyme adds the modifying group, and another can act to remove it. For example, proteins are phosphorylated by enzymes known as protein kinases, while protein phosphatases remove phosphate groups. Post-translational modifications act in much the same way as do allosteric effectors, they modify the structure and, in turn, the activity of the polypeptide to which they are attached. They can also modify a proteins interactions with other proteins, the protein's localization within the cell, or its stability. |
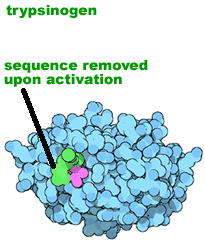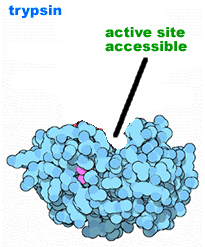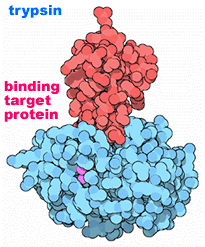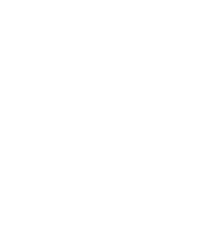 |
Proteolytic processing: Another method for regulating protein activity involves the cleavage of the polypeptide chain. Many proteins are originally synthesized in a longer, and inactive "pro-form". To become active the pro-peptide must be removed - it is cut by a protease. This proteolytic processing activates the protein. Proteolytic processing is itself often regulated. |
|
|
|
Questions to answer
|
|
|
Questions to ponder
|
|
|
|
| replace with revised beSocratic activity |
|
|
|
Tweet
last revision
10-May-2014
|